Note
Click here to download the full example code
Field interpolation example using equivalent surface currents¶
import numpy as np
from bfieldtools.mesh_conductor import MeshConductor, StreamFunction
from mayavi import mlab
import trimesh
import mne
PLOT = True
SAVE_FIGURES = True
IMPORT_MNE_DATA = True
SAVE_MNE_DATA = True
SAVE_DIR = "./MNE interpolation/"
from pyface.api import GUI
_gui = GUI()
First, let’s import the MEG data
if IMPORT_MNE_DATA:
from mne.datasets import sample
data_path = sample.data_path()
fname = data_path + "/MEG/sample/sample_audvis-ave.fif"
# Reading
condition = "Left Auditory"
evoked = mne.read_evokeds(fname, condition=condition, baseline=(None, 0), proj=True)
evoked.pick_types(meg="mag")
# evoked.plot(exclude=[], time_unit="s")
i0, i1 = evoked.time_as_index(0.08)[0], evoked.time_as_index(0.09)[0]
field = evoked.data[:, i0:i1].mean(axis=1)
# Read BEM for surface geometry and transform to correct coordinate system
import os.path as op
subject = "sample"
subjects_dir = op.join(data_path, "subjects")
bem_fname = op.join(
subjects_dir, subject, "bem", subject + "-5120-5120-5120-bem-sol.fif"
)
bem = mne.read_bem_solution(bem_fname)
# Head mesh 0
# Innerskull mesh 2
surf_index = 2
trans_fname = op.join(data_path, "MEG", "sample", "sample_audvis_raw-trans.fif")
trans0 = mne.read_trans(trans_fname)
R = trans0["trans"][:3, :3]
t = trans0["trans"][:3, 3]
# Surface from MRI to HEAD
rr = (bem["surfs"][surf_index]["rr"] - t) @ R
# Surface from HEAD to DEVICE
trans1 = evoked.info["dev_head_t"]
R = trans1["trans"][:3, :3]
t = trans1["trans"][:3, 3]
rr = (rr - t) @ R
innerskull = trimesh.Trimesh(rr, bem["surfs"][surf_index]["tris"])
surf_index = 0
R = trans0["trans"][:3, :3]
t = trans0["trans"][:3, 3]
# Surface from MRI to HEAD
rr = (bem["surfs"][surf_index]["rr"] - t) @ R
# Surface from HEAD to DEVICE
R = trans1["trans"][:3, :3]
t = trans1["trans"][:3, 3]
rr = (rr - t) @ R
head = trimesh.Trimesh(rr, bem["surfs"][surf_index]["tris"])
mesh = head
# Sensor locations and directions in DEVICE coordinate system
p = np.array(
[
ch["loc"][:3]
for ch in evoked.info["chs"]
if ch["ch_name"][-1] == "1" and ch["ch_name"][:3] == "MEG"
]
)
n = np.array(
[
ch["loc"][-3:]
for ch in evoked.info["chs"]
if ch["ch_name"][-1] == "1" and ch["ch_name"][:3] == "MEG"
]
)
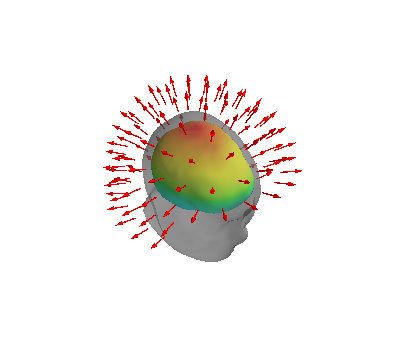
if PLOT:
# Plot sensor locations and directions
fig = mlab.figure(bgcolor=(1, 1, 1))
mlab.triangular_mesh(*innerskull.vertices.T, innerskull.faces)
mlab.triangular_mesh(
*head.vertices.T, head.faces, color=(0.5, 0.5, 0.5), opacity=0.5
)
mlab.quiver3d(*p.T, *n.T, mode="arrow")
fig.scene.isometric_view()
if SAVE_FIGURES:
mlab.savefig(SAVE_DIR + "MEG_geometry.png", magnification=4, figure=fig)
if SAVE_MNE_DATA:
np.savez(
SAVE_DIR + "mne_data.npz",
mesh=head,
p=p,
n=n,
vertices=head.vertices,
faces=head.faces,
)
evoked.save(SAVE_DIR + "left_auditory-ave.fif")
else:
with np.load(SAVE_DIR + "mne_data.npz", allow_pickle=True) as data:
mesh = data["mesh"]
p = data["p"]
n = data["n"]
mesh = trimesh.Trimesh(vertices=data["vertices"], faces=data["faces"])
evoked = mne.Evoked(SAVE_DIR + "left_auditory-ave.fif")
Out:
Reading /home/rzetter/mne_data/MNE-sample-data/MEG/sample/sample_audvis-ave.fif ...
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Found the data of interest:
t = -199.80 ... 499.49 ms (Left Auditory)
0 CTF compensation matrices available
nave = 55 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
Applying baseline correction (mode: mean)
Loading surfaces...
Three-layer model surfaces loaded.
Loading the solution matrix...
Loaded linear_collocation BEM solution from /home/rzetter/mne_data/MNE-sample-data/subjects/sample/bem/sample-5120-5120-5120-bem-sol.fif
Fit the surface current for the auditory evoked response
c = MeshConductor(mesh_obj=mesh, basis_name="suh", N_suh=150)
M = c.mass
sensor_coupling = np.einsum("ijk,ij->ik", c.B_coupling(p), n)
# a = np.linalg.pinv(sensor_coupling, rcond=1e-15) @ field
ss = np.linalg.svd(sensor_coupling @ sensor_coupling.T, False, False)
# reg_exps = [0.5, 1, 2, 3, 4, 5, 6, 7, 8]
reg_exps = [1]
rel_errors = []
for reg_exp in reg_exps:
_lambda = np.max(ss) * (10 ** (-reg_exp))
# Laplacian in the suh basis is diagonal
BB = sensor_coupling.T @ sensor_coupling + _lambda * (-c.laplacian) / np.max(
abs(c.laplacian)
)
a = np.linalg.solve(BB, sensor_coupling.T @ field)
s = StreamFunction(a, c)
b_filt = sensor_coupling @ s
rel_error = np.linalg.norm(b_filt - field) / np.linalg.norm(field)
print("Relative error:", rel_error * 100, "%")
rel_errors.append(rel_error)
if PLOT:
fig = mlab.figure(bgcolor=(1, 1, 1))
surf = s.plot(False, figure=fig)
surf.actor.mapper.interpolate_scalars_before_mapping = True
surf.module_manager.scalar_lut_manager.number_of_colors = 16
if SAVE_FIGURES:
mlab.savefig(
SAVE_DIR + "SUH_scalp_streamfunction.png", magnification=4, figure=fig
)

Out:
Calculating surface harmonics expansion...
Computing the laplacian matrix...
Computing the mass matrix...
Closed mesh or Neumann BC, leaving out the constant component
Computing the mass matrix...
Computing magnetic field coupling matrix, 2562 vertices by 102 target points... took 0.23 seconds.
Computing the laplacian matrix...
Relative error: 2.0549237013162216 %
Interpolate MEG data to the sensor surface
from bfieldtools.utils import load_example_mesh
helmet = load_example_mesh("meg_helmet", process=False)
# Bring the surface roughly to the correct place
helmet.vertices[:, 2] -= 0.05
# Reset coupling by hand
c.B_coupling.reset()
mlab.figure(bgcolor=(1, 1, 1))
B_surf = np.sum(
c.B_coupling(helmet.vertices) * helmet.vertex_normals[:, :, None], axis=1
)
if PLOT:
fig = mlab.quiver3d(*p.T, *n.T, mode="arrow")
scalars = B_surf @ s
surf = mlab.triangular_mesh(
*helmet.vertices.T, helmet.faces, scalars=scalars, colormap="seismic"
)
surf.actor.mapper.interpolate_scalars_before_mapping = True
surf.module_manager.scalar_lut_manager.number_of_colors = 15
if SAVE_FIGURES:
mlab.savefig(
SAVE_DIR + "SUH_sensors_streamfunction.png", magnification=4, figure=fig
)
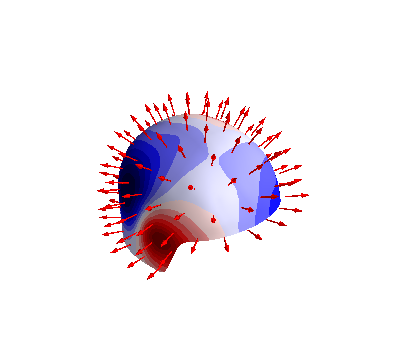
Out:
Computing magnetic field coupling matrix, 2562 vertices by 2044 target points... took 1.36 seconds.
Calculate magnetic field in volumetric grid
Nvol = 30
x = np.linspace(-0.125, 0.125, Nvol)
vol_points = np.array(np.meshgrid(x, x, x, indexing="ij")).reshape(3, -1).T
# mlab.points3d(*vol_points.T)
c.B_coupling.reset()
Bvol_coupling = c.B_coupling(vol_points, Nchunks=100, analytic=True)
s = StreamFunction(a, c)
# s = StreamFunction(a, c)
Bvol = Bvol_coupling @ s
Out:
Computing magnetic field coupling matrix analytically, 2562 vertices by 27000 target points... took 82.12 seconds.
Now, plot the computed magnetic field using streamlines
if PLOT:
from bfieldtools.mesh_calculus import gradient
fig = mlab.figure(bgcolor=(1, 1, 1))
surf = s.plot(False, figure=fig)
surf.actor.mapper.interpolate_scalars_before_mapping = True
surf.module_manager.scalar_lut_manager.number_of_colors = 16
vecs = mlab.pipeline.vector_field(
*vol_points.T.reshape(3, Nvol, Nvol, Nvol), *Bvol.T.reshape(3, Nvol, Nvol, Nvol)
)
vecnorm = mlab.pipeline.extract_vector_norm(vecs)
seed_points = mesh.vertices[mesh.faces].mean(axis=1) - 0.01 * mesh.face_normals
seed_vals = c.basis @ c.inductance @ s
seed_vals_grad = np.linalg.norm(gradient(seed_vals, c.mesh), axis=0)
seed_vals = abs(seed_vals[mesh.faces].mean(axis=1)) ** 2
seed_vals[seed_vals_grad > seed_vals_grad.max() / 1.8] = 0
Npoints = 500
seed_inds = np.random.choice(
np.arange(len(seed_vals)), Npoints, False, seed_vals / seed_vals.sum()
)
seed_points = seed_points[seed_inds]
streams = []
for pi in seed_points:
streamline = mlab.pipeline.streamline(
vecnorm,
integration_direction="both",
colormap="BuGn",
seed_visible=False,
seedtype="point",
)
streamline.seed.widget.position = pi
streamline.stream_tracer.terminal_speed = 3e-13
streamline.stream_tracer.maximum_propagation = 0.1
streamline.actor.property.render_lines_as_tubes = True
streamline.actor.property.line_width = 4.0
streams.append(streamline)
# Custom colormap with alpha channel
streamine = streams[0]
lut = streamline.module_manager.scalar_lut_manager.lut.table.to_array()
lut[:, -1] = np.linspace(0, 255, 256)
streamline.module_manager.scalar_lut_manager.lut.table = lut
streamline.module_manager.scalar_lut_manager.data_range = np.array(
[1.0e-13, 1.0e-12]
)
for streamline in streams:
streamline.stream_tracer.terminal_speed = 1e-13
streamline.seed.widget.hot_spot_size = 0.1
streamline.stream_tracer.initial_integration_step = 0.01
streamline.stream_tracer.minimum_integration_step = 0.1
sensors = mlab.quiver3d(*p.T, *n.T, mode="cylinder")
sensors.glyph.glyph_source.glyph_source.height = 0.1
sensors.actor.property.color = (0.5, 0.5, 0.5)
sensors.actor.mapper.scalar_visibility = False
sensors.glyph.glyph_source.glyph_source.resolution = 32
sensors.glyph.glyph.scale_factor = 0.03
# sensors.glyph.glyph_source.glyph_source.shaft_radius = 0.05
fig.scene.camera.position = [
0.637392177469018,
0.07644693029292644,
-0.07183513804689762,
]
fig.scene.camera.focal_point = [
-6.413459777832031e-05,
0.01716560870409012,
-0.0229007127850005,
]
fig.scene.camera.view_angle = 30.0
fig.scene.camera.view_up = [
0.04390624852005244,
0.3114421192517664,
0.9492502555685007,
]
fig.scene.camera.clipping_range = [0.3366362817578398, 1.0281065506557443]
fig.scene.camera.compute_view_plane_normal()
while fig.scene.light_manager is None:
_gui.process_events()
camera_light = fig.scene.light_manager.lights[0]
camera_light.intensity = 0.7
if SAVE_FIGURES:
mlab.savefig(
SAVE_DIR + "SUH_streamlines_lateral.png", figure=fig, magnification=4
)
fig.scene.camera.position = [
-6.413459777832031e-05,
0.01716560870409012,
0.6191735842078244,
]
fig.scene.camera.focal_point = [
-6.413459777832031e-05,
0.01716560870409012,
-0.0229007127850005,
]
fig.scene.camera.view_angle = 30.0
fig.scene.camera.view_up = [0.0, 1.0, 0.0]
fig.scene.camera.clipping_range = [0.3381552363433513, 1.0261944997830243]
fig.scene.camera.compute_view_plane_normal()
if SAVE_FIGURES:
mlab.savefig(
SAVE_DIR + "SUH_streamlines_coronal.png", figure=fig, magnification=4
)
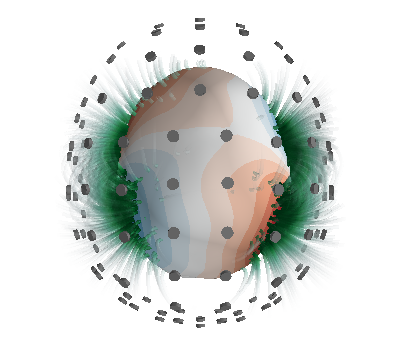
Out:
Computing the inductance matrix...
Computing self-inductance matrix using rough quadrature (degree=2). For higher accuracy, set quad_degree to 4 or more.
Estimating 24058 MiB required for 2562 by 2562 vertices...
Computing inductance matrix in 80 chunks (7035 MiB memory free), when approx_far=True using more chunks is faster...
Computing triangle-coupling matrix
Inductance matrix computation took 10.99 seconds.
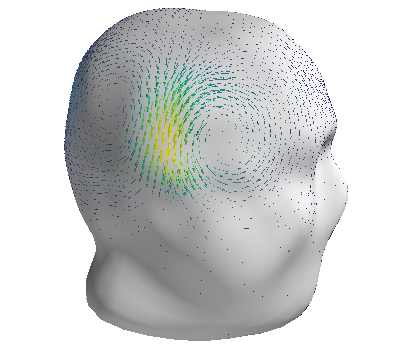
Finally, plot the surface current density itself
fig = mlab.figure(bgcolor=(1, 1, 1))
grad_s = gradient(c.basis @ s, mesh, rotated=True)
q = mlab.quiver3d(
*(mesh.vertices[mesh.faces].mean(axis=1).T),
*grad_s,
colormap="viridis",
mode="arrow"
)
mlab.triangular_mesh(*head.vertices.T, head.faces, color=(0.8, 0.8, 0.8), opacity=1.0)
fig.scene.camera.position = [
0.4987072212753703,
0.06469079487766746,
-0.0014732384935239248,
]
fig.scene.camera.focal_point = [
0.0018187984824180603,
0.012344694641686624,
-0.04367139294433087,
]
fig.scene.camera.view_angle = 30.0
fig.scene.camera.view_up = [
-0.10720122151366927,
0.23975383168819672,
0.9648968848000314,
]
fig.scene.camera.clipping_range = [0.28329092545021717, 0.7772019991936254]
fig.scene.camera.compute_view_plane_normal()
if SAVE_FIGURES:
mlab.savefig(
SAVE_DIR + "SUH_surface_currents_lateral.png", figure=fig, magnification=4
)
Total running time of the script: ( 6 minutes 11.527 seconds)
Estimated memory usage: 8527 MB
